GPN (基因组预训练网络)
[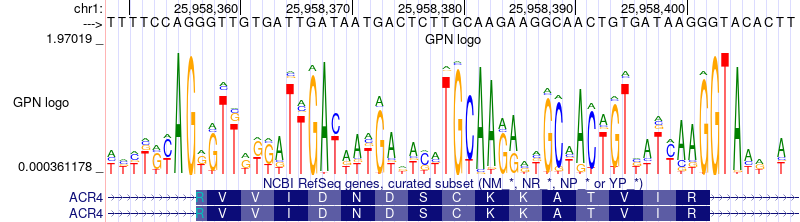 ](https://raw.githubusercontent.com/songlab-cal/gpn/main/ https://genome.ucsc.edu/s/gbenegas/gpn-arabidopsis)
](https://raw.githubusercontent.com/songlab-cal/gpn/main/ https://genome.ucsc.edu/s/gbenegas/gpn-arabidopsis)
目录
安装
pip install git+https://github.com/songlab-cal/gpn.git
最小使用示例
import gpn.model
from transformers import AutoModelForMaskedLM
model = AutoModelForMaskedLM.from_pretrained("songlab/gpn-brassicales")
# 或
model = AutoModelForMaskedLM.from_pretrained("songlab/gpn-msa-sapiens")
GPN
也可称为GPN-SS(单序列)。
示例
特定论文的代码和资源
使用自己的数据进行训练
- [创建数据集的Snakemake工作流程](https://github.com/songlab-cal/gpn/blob/main/workflow/make_dataset
- 可以根据给定的登录号列表自动从NCBI下载数据,或使用您自己的fasta文件。
- 训练
- 将自动检测所有可用的GPU。
- 在Weights & Biases上跟踪指标。
- 已实现的模型:
ConvNet,GPNRoFormer(Transformer) - 指定配置覆盖:例如
--config_overrides n_layers=30 - 示例:
WANDB_PROJECT=your_project torchrun --nproc_per_node=$(echo $CUDA_VISIBLE_DEVICES | awk -F',' '{print NF}') -m gpn.ss.run_mlm --do_train --do_eval \
--fp16 --report_to wandb --prediction_loss_only True --remove_unused_columns False \
--dataset_name results/dataset --tokenizer_name gonzalobenegas/tokenizer-dna-mlm \
--soft_masked_loss_weight_train 0.1 --soft_masked_loss_weight_evaluation 0.0 \
--weight_decay 0.01 --optim adamw_torch \
--dataloader_num_workers 16 --seed 42 \
--save_strategy steps --save_steps 10000 --evaluation_strategy steps \
--eval_steps 10000 --logging_steps 10000 --max_steps 120000 --warmup_steps 1000 \
--learning_rate 1e-3 --lr_scheduler_type constant_with_warmup \
--run_name your_run --output_dir your_output_dir --model_type ConvNet \
--per_device_train_batch_size 512 --per_device_eval_batch_size 512 --gradient_accumulation_steps 1 \
--torch_compile
- 提取嵌入
- 输入文件需要包含
chrom、start、end - 示例:
- 输入文件需要包含
torchrun --nproc_per_node=$(echo $CUDA_VISIBLE_DEVICES | awk -F',' '{print NF}') -m gpn.ss.get_embeddings windows.parquet genome.fa.gz 100 your_output_dir \
results.parquet --per-device-batch-size 4000 --is-file --dataloader-num-workers 16
- 变异效应预测
- 输入文件需要包含
chrom、pos、ref、alt - 示例:
- 输入文件需要包含
torchrun --nproc_per_node=$(echo $CUDA_VISIBLE_DEVICES | awk -F',' '{print NF}') -m gpn.ss.run_vep variants.parquet genome.fa.gz 512 your_output_dir results.parquet \
--per-device-batch-size 4000 --is-file --dataloader-num-workers 16
GPN-MSA
示例
- 模型使用示例:
examples/msa/basic_example.ipynb - 变异效应预测:
examples/msa/vep.ipynb - 训练(人类):
examples/msa/training.ipynb
特定论文的代码和资源
在其他物种上训练(如植物)
正在建设中。
引用
GPN:
@article{benegas2023dna,
author = {Gonzalo Benegas and Sanjit Singh Batra and Yun S. Song },
title = {DNA language models are powerful predictors of genome-wide variant effects},
journal = {Proceedings of the National Academy of Sciences},
volume = {120},
number = {44},
pages = {e2311219120},
year = {2023},
doi = {10.1073/pnas.2311219120},
URL = {https://www.pnas.org/doi/abs/10.1073/pnas.2311219120},
eprint = {https://www.pnas.org/doi/pdf/10.1073/pnas.2311219120},
}
GPN-MSA:
@article{benegas2023gpnmsa,
author = {Gonzalo Benegas and Carlos Albors and Alan J. Aw and Chengzhong Ye and Yun S. Song},
title = {GPN-MSA: an alignment-based DNA language model for genome-wide variant effect prediction},
elocation-id = {2023.10.10.561776},
year = {2023},
doi = {10.1101/2023.10.10.561776},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2023/10/11/2023.10.10.561776},
eprint = {https://www.biorxiv.org/content/early/2023/10/11/2023.10.10.561776.full.pdf},
journal = {bioRxiv}
}

 Github
Github 论文
论文










